Thank you to everyone who completed the 2024 gnomAD user survey. Your feedback is invaluable in helping us improve gnomAD for all users. In this blog post, we discuss the feedback we received, actions taken or planned to address requests or pain points, and any issues that remain unresolvable.
2024 User Survey Feedback
We sought feedback for three main areas in this survey: prioritization of features not yet in gnomAD v4, requests for new features or modifications of existing features, and description of pain points. The sections below summarize the 325 responses to our survey.
Prioritization of features not yet in gnomAD v4
gnomAD v4.0 was a minimum viable product (MVP) release, which meant that it did not include some of the features included in previous versions. We asked our users to rank the v4.0 non-MVP features to assess their value within our user community:
- GTEx data and pext scores
- Genetic ancestry sub-group information
- Multinucleotide variant (MNV) calls
- Variant co-occurrence (statistical phasing and per-gene counts)
- Linkage disequilibrium scores
- Regional missense constraint and MPC scores
- Pharmacogenomic (PGx) named alleles
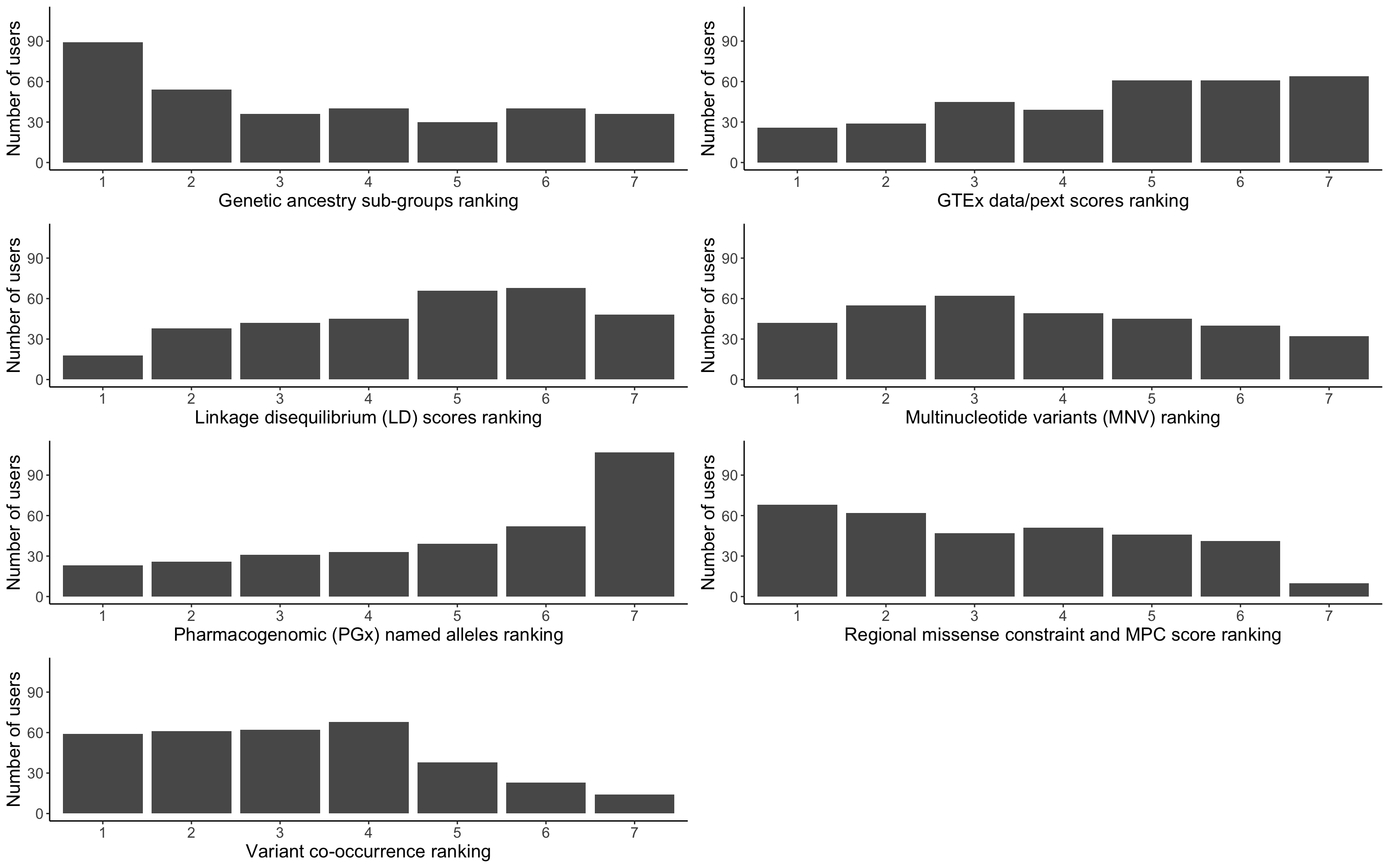
The prioritized list of features based on mean user ranking is:
- Variant co-occurrence (statistical phasing and per-gene counts)
- Regional missense constraint and MPC scores
- Genetic ancestry sub-group information
- Multinucleotide variant (MNV) calls
- Linkage disequilibrium scores
- GTEx data and pext scores
- Pharmacogenomic (PGx) named alleles
Requested enhancements
User requests for new features or modifications of current gnomAD features generally fell into one of the following categories:
- Variant search-related requests: Users expressed the desire to streamline variant searching by implementing a more flexible variant search option (e.g., allowing searches on HGVS nomenclature) or support for RefSeq transcripts.
- Phenotype-related requests: Users expressed interest in knowing the cohort of origin for samples or phenotype information where available.
- Dataset subsets: Users expressed interest in stratifying gnomAD v4 using non-disease subsets.
- Re-implementation of gnomAD v4.0 non-MVP feature requests: Users requested re-implementation of the features prioritized above.
- New data or feature requests: Users requested more data (e.g., increased sample sizes; more in silico prediction scores) and new features (e.g., more links out to external resources; displaying exon numbers).
- Documentation requests: Users requested tutorial videos and additional documentation to maximize usage of the gnomAD resource.
Pain points
The reported pain points fell into three main categories:
- Browser performance: Users reported issues loading the gnomAD browser or slow loading of pages.
- Downloads: Users reported issues with the downloadable file size and the inability to download specific data without downloading large files.
- Perception that v4 is not as healthy as previous datasets: Users expressed concern at the increased inclusion of biobank samples in v4.
Addressing User Feedback: Actions Taken and Planned
The feedback received in the user survey directly informed several key actions and planned initiatives. The team prioritized addressing the pain points reported in the survey and have implemented new browser security measures and two new features to facilitate programmatic gnomAD data access: the gnomAD API and the gnomAD toolbox.
Browser performance updates
Over 11% of survey respondents reported experiencing slow loading times on the browser. We analyzed overall app traffic and found that roughly half of it was generated by known bots and scrapers since the end of last year. To remediate this, we have implemented additional protective measures against web scrapers and other agents that downgrade browser performance. After implementing targeted protections, we were able to block this traffic to improve browser performance for users. As a result, the monthly rate of slow performance warnings dropped by roughly 90%.
Enabling easier data access
The size of gnomAD downloadable files increases with each release. To enable continued programmatic access to gnomAD and the downloads, we have added two new features: support for the gnomAD API and a new utility repository named the gnomAD toolbox. Both the API and the gnomAD toolbox allow users to query gnomAD data without requiring any data downloads.
Note that due to security and performance reasons, users are limited to 10 requests per IP address per 60-second period. The gnomAD toolbox does not have usage limits and is not tied to cloud computing; for more information, please see our previous blog post.
Work in progress
We are actively working on addressing user requests for new features and modifications to existing gnomAD functionalities.
More data
The gnomAD production team is actively developing the next major release, v5, which will substantially expand the resource by incorporating the All of Us v8 release (414,830 genomes). The inclusion of All of Us data will enhance ancestral representation within gnomAD and introduce new structural variation. It will enable users with access to All of Us data to link genotype and phenotype information. Furthermore, in response to user feedback, we will incorporate AlphaMissense scores into v5. We anticipate that v5 will be released in 2026.
Non-MVP feature re-implementation
The gnomAD team is a small team with limited bandwidth, and our top priority is developing the next gnomAD version. However, we are also working to restore features from previous versions that are not yet available in gnomAD v4. As a part of this effort, we released GTEx data and pext scores on genome reference build GRCh38 in November 2024. We are also actively regenerating gnomAD regional missense constraint and MPC metrics, which are expected to be released this fall.
We are also planning to regenerate variant co-occurrence and multinucleotide variant calls but do not have anticipated release dates for these features yet.
Browser updates
A few new browser features that are actively being implemented are:
- Inclusion of ClinVar variants track on copy number variant and structural variant gene pages
- Inclusion of exon numbers and exon start and end coordinates in the transcript track
- Inclusion of HGVS coding sequence consequence on variant pages
- Updating the behavior of control+f on gene pages to search all variants in a gene, not just currently displayed variants
These features will be implemented on a rolling basis, and we anticipate they will be completed by the end of this year.
Unresolvable issues
There are a few points of user feedback we are unable to address.
Non-MVP feature re-implementation
One of the top ranked features from the user survey was re-implementation of genetic ancestry subgroup information. However, we do not have plans to regenerate genetic ancestry subgroups at this time. Genetic ancestry groupings have been misappropriated with malicious intent1, and we do not believe the additional subgroups we can identify with our limited sample metadata would be valuable for clinical variant classification or risk assessment. This is an area of active community discussion.
In addition, due to cost constraints, we will not be calculating linkage disequilibrium (LD) scores or generating pharmacogenomic haplotypes. Users needing these metrics should apply for access to the All of Us (AoU) dataset. Pharmacogenomic data are available through All of Us, and the gnomAD team is currently collaborating on an effort to generate LD scores from the AoU genomes.
Phenotype information
The gnomAD resource does not have permission, access, or staffing to investigate the availability of phenotype data for individual samples nor the consent status to share that data. We are also unable to provide any information about the clinical status of a sample or their cohort or project of origin. However, in a future release, it is our plan to be able to note that a sample is from a cohort for which users can apply for access (UK Biobank, AoU, etc).
Disease subsets and perception that v4 is not as healthy as previous datasets
In the past, the gnomAD browser has supported disease-specific subsets (e.g., non-cancer). However, we have removed support for disease-specific subsets for two reasons: sample metadata and sample size. While we had high-level study phenotype and case/control status for some samples, we do not have comprehensive phenotype metadata for most gnomAD samples, and many samples are now derived from large biobanks, which can include individuals with disease. As such, we cannot ensure that samples in a non-disease subset do not have the specified disease.
Additionally, as the dataset has grown, concerns about enrichment of any particular phenotype has decreased. We continue to remove cohorts recruited for severe pediatric disease, except for a small number of diverse cohorts where we have included unaffected relatives. We have also removed the TCGA cancer samples due to data quality. For a more detailed understanding of the cohorts included in gnomAD, please see the list shared on our About page or the “Study Diseases in gnomAD” table on our Stats page.
Liftover
We will not be releasing a GRCh37 liftover version of gnomAD v4. The liftover feature in the browser is designed for mapping variants between different gnomAD versions, and we do not intend to update it to display liftover coordinates for variants found in only a single human genome reference version.
Variant search
We received feedback from several users requesting more flexible variant search nomenclature in the browser (e.g., enabling search for variants using HGVS consequence or RefSeq transcript IDs). We recognize that implementing support for different nomenclatures is important, but this would require significant changes to the search logic and possibly a complete redesign, that our team does not have bandwidth to complete at this time.
Contact us
Your feedback is important to us! gnomAD is always improving, and we appreciate community input.
Here’s how you can connect with us:
- Virtual Feedback & Office Hours: We held our first session in March and invite you to email gnomad@broadinstitute.org if you’d like to participate next year.
- Forum: For ongoing feedback, questions, or feature requests, please post on our forum.
Thank you for helping us improve gnomAD.
References
- Carlson, J., Henn, B. M., Al-Hindi, D. R., & Ramachandran, S. (2022). Counter the weaponization of genetics research by extremists. Nature, 610(7932), 444–447. https://doi.org/10.1038/d41586-022-03252-z