We’re excited to announce the addition of mitochondrial genome (mtDNA) constraint metrics to gnomAD, developed using data from over 56,000 individuals in gnomAD v4.11,2. These new metrics are designed to identify regions of the mtDNA under strong selective pressure that are thus most likely to harbor functionally important and disease-associated variants. The gnomAD browser now features mitochondrial gene constraint and regional constraint metrics.
Background
Constraint metrics for the nuclear genome have been valuable tools for identifying deleterious genetic variation underlying disease. However, the constraint model and metrics for the mitochondrial and nuclear genome differ due to the unique characteristics of the mtDNA, which includes its smaller size, lack of introns, high copy number, presence of heteroplasmy, distinct mutational mechanisms, and higher rate of mutation. To address this, we developed a constraint model specifically for the mitochondrial genome and applied it to the dataset of mtDNA variation in gnomAD v4.1 to compute mitochondrial constraint metrics. Key differences between the mitochondrial and nuclear constraint models include:
- Mutational Model: A mitochondrial mutational model was developed and applied, as nuclear mutational models could not be used due to distinct mutational mechanisms and signatures between the genomes.
- Observed/Expected Calculation: Nuclear constraint models assess the number of unique variants, while mitochondrial constraint models evaluate the sum of maximum heteroplasmy for variants.
- Confidence Interval: A beta distribution, rather than a Poisson distribution, is used to calculate confidence intervals for the observed/expected ratios based on maximum heteroplasmy values.
Methods
We calculated constraint metrics for all protein-coding, rRNA, and tRNA genes in the mitochondrial genome by comparing observed variation in gnomAD to expected levels under neutrality. The observed value is the sum of maximum heteroplasmy levels for all SNVs in a gene, while the expected sum maximum heteroplasmy value is computed using a mitochondrial mutational model accounting for trinucleotide context. Heteroplasmy – the proportion of mtDNA copies carrying a variant – is important to account for since most pathogenic variants have low heteroplasmy in gnomAD, reflecting that individuals can be asymptomatic if heteroplasmy is low enough. The observed/expected (oe) ratio provides an inference on the strength of selection against gene variation, with a 90% confidence interval to capture uncertainty around the ratio.
Regional constraint identifies regions within genes that are more constrained than the entire gene. We provide regional constraint metrics for all protein and rRNA genes in the mtDNA. For regional missense constraint in protein genes, the missense oe ratio was calculated for all regions ≥30 bp. Regions with significantly lower oe ratios than the gene’s oe ratio were identified using a beta distribution, and a greedy algorithm prioritized the most significant, non-overlapping intervals. The false discovery rate was estimated using random permutations, with only high-confidence intervals retained. Regional constraint in rRNA genes was evaluated similarly. Note regional constraint metrics are not provided for tRNA genes due to their small size.
To learn more about the mitochondrial genome constraint model, see Lake et al. Nature 2024.
Mitochondrial Gene Constraint Metrics
Gene constraint metrics are now displayed on the top right hand corner of the gene landing page in the browser, akin to how they are displayed for nuclear genes. Metrics are provided for synonymous, missense, and stop-gain variants in protein-coding genes (Figure 1), and for all SNVs in RNA genes (Figure 2).
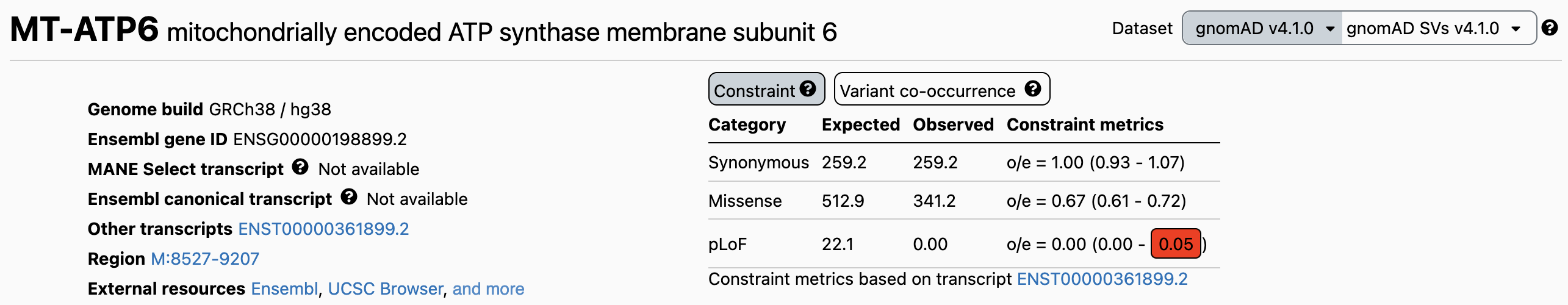
Figure 1: Example of gene constraint metrics in the browser for a protein-coding gene.
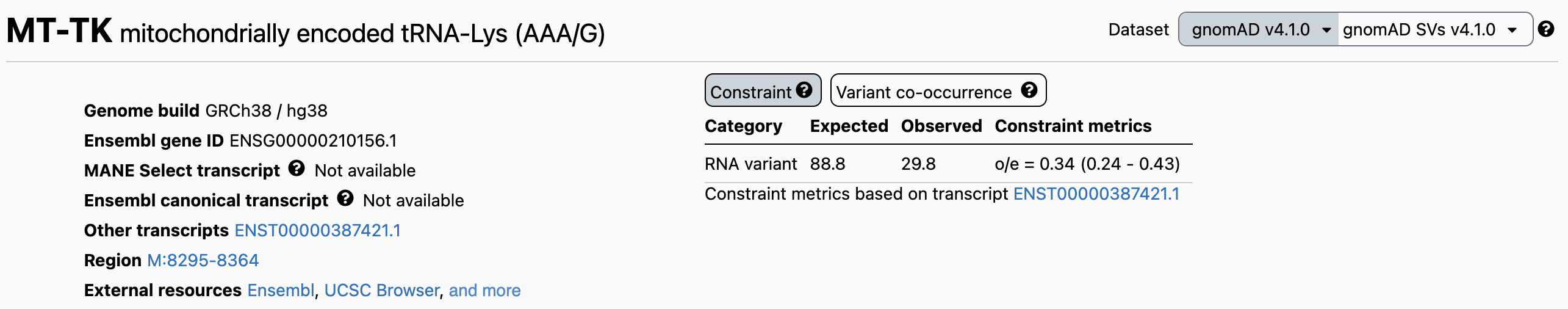
Figure 2: Example of gene constraint metrics in the browser for a RNA gene.
The constraint table shows the expected and observed sum maximum heteroplasmy, their oe ratio, and 90% confidence interval (CI). We recommend using the upper bound of the CI, the OEUF (observed to expected upper bound fraction), as a conservative measure of constraint. A lower OEUF indicates stronger selection, while a higher value suggests greater tolerance.
More detailed information on the mitochondrial gene constraint metrics can be found here.
Mitochondrial Gene Regional Constraint Metrics
Regional constraint metrics are visible on the gene landing page under the transcript track in the browser (Figure 3).
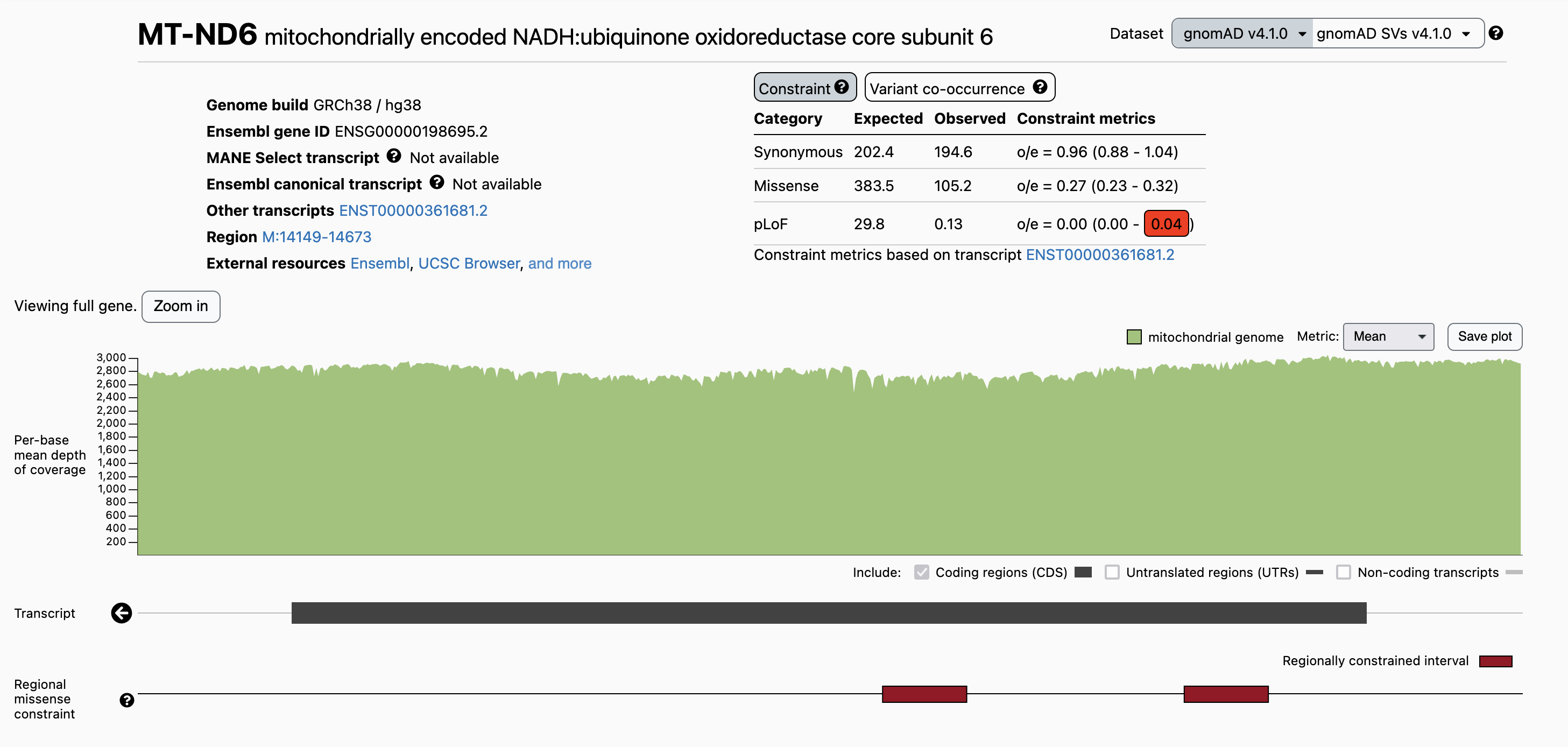
Figure 3: Example of regional missense constraint in the browser for a protein-coding gene.
The regional constraint track displays regionally constrained intervals in the gene in maroon. Hovering over an interval will reveal its coordinates, oe ratio, and 90% CI (Figure 3). These regionally constrained intervals are enriched for pathogenic variants and functionally important residues1. Knowing if a variant falls within an interval of regional constraint can thus help prioritize variants most likely to have a deleterious functional impact.
More detailed information on the mitochondrial regional constraint metrics can be found here.
Summary
In summary, we have added constraint metrics for the mitochondrial genome to the gnomAD browser. We hope these metrics will aid clinicians and researchers in prioritizing mitochondrial genome variants found in individuals with disease. To learn more about mitochondrial genome constraint metrics, including a more detailed overview, please see Lake et al. Nature 2024.
The mitochondrial genome constraint metrics are also available for download. For additional questions about these metrics, please contact Nicole Lake.
Acknowledgments
With thanks to the gnomAD team for providing valuable feedback on the mitochondrial genome constraint metrics. Specifically, we are grateful to Katherine Chao and Phil Darnowsky for their guidance and work integrating the mitochondrial genome constraint metrics into the gnomAD browser. We also thank the PIs, researchers, and participants of each study that contributed data to gnomAD that enabled the generation of these constraint metrics.
References:
- Lake NJ, Ma K, Liu W, Battle SL, Laricchia KM, Tiao G, Puiu D, Ng KK, Cohen J, Compton AG, Cowie S, Christodoulou J, Thorburn DR, Zhao H, Arking DE, Sunyaev SR, Lek M. (2024). Quantifying constraint in the human mitochondrial genome. Nature. Nov;635(8038):390-397. https://doi.org/10.1038/s41586-024-08048-x
- Laricchia KM*, Lake NJ*, Watts NA, Shand M, Haessly A, Gauthier L, Benjamin D, Banks E, Soto J, Garimella K, Emery J; Genome Aggregation Database Consortium; Rehm HL, MacArthur DG, Tiao G#, Lek M#, Mootha VK#, Calvo SE#. (2022). Mitochondrial DNA variation across 56,434 individuals in gnomAD. Genome Research. Mar;32(3):569-582. https://doi.org/10.1101/gr.276013.121. *Contributed equally. #Co-senior authors.