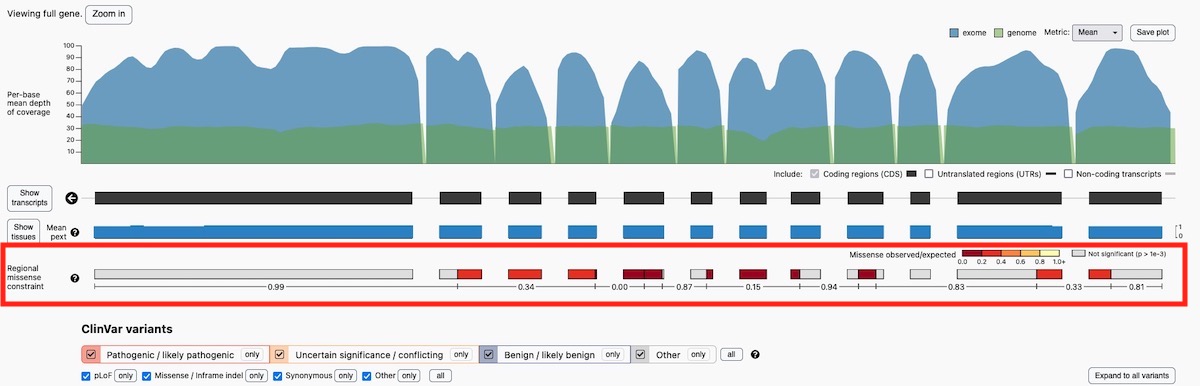
The regional missense constraint (RMC) track has now been added to the gene pages in the gnomAD v2 browser.
This track was previously only available on the ExAC browser and has been updated to include three distinct views:
- Genes that exhibit evidence of regional variability in missense intolerance will have regional missense constraint information displayed
- Genes that were searched for but that did not exhibit any evidence of regional differences in missense intolerance will have transcript-wide missense constraint information displayed
- Outlier genes will have text indicating that these genes were not searched for regional differences in missense intolerance
Note that regional missense constraint was only calculated on canonical transcripts (single canonical transcript per gene).
The updated track has a new color scheme to more clearly visualize regions of missense intolerance and a more detailed hover-over to provide more information about each domain of regional missense constraint.
For more information about how these data were generated, please see the RMC help page.