To help identify biologically relevant transcripts, the gene page includes information from the GTEx project on transcripts’ expression in different tissues.

Previously, this was displayed next to transcripts in the transcripts track. However, only one tissue could be selected at a time and the x-axis was scaled based on the selected tissue, making it difficult to compare expression across tissues.

Now, the transcripts track shows a dot next to each transcript, the size of which is scaled based on that transcript’s mean expression across all tissues. This maintains the ability to find the most expressed transcript(s) at a glance. Hovering the pointer over the dot shows the transcript’s mean expression across all tissues as well as the tissue in which the transcript is most expressed.
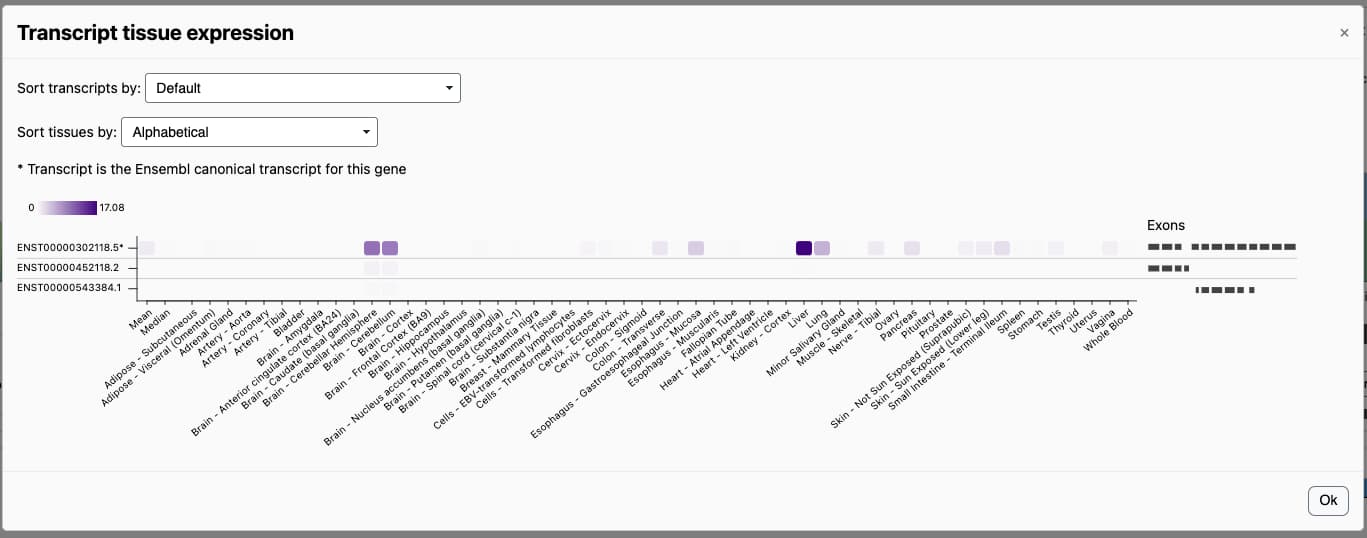
Information on expression in specific tissues can be seen by pressing the “Show transcript tissue expression” button. This brings up a heatmap that displays expression for all transcripts in all tissues.