Transcript tissue expression information from the GTEx project has been added to the individual tissue pext tracks.
Expanding the pext track on the gene page shows which exonic regions are expressed in each tissue. However, since the y-axis of each track is normalized 0 to 1, it does not show which tissues the gene is expressed in. That information is available in the transcripts track.
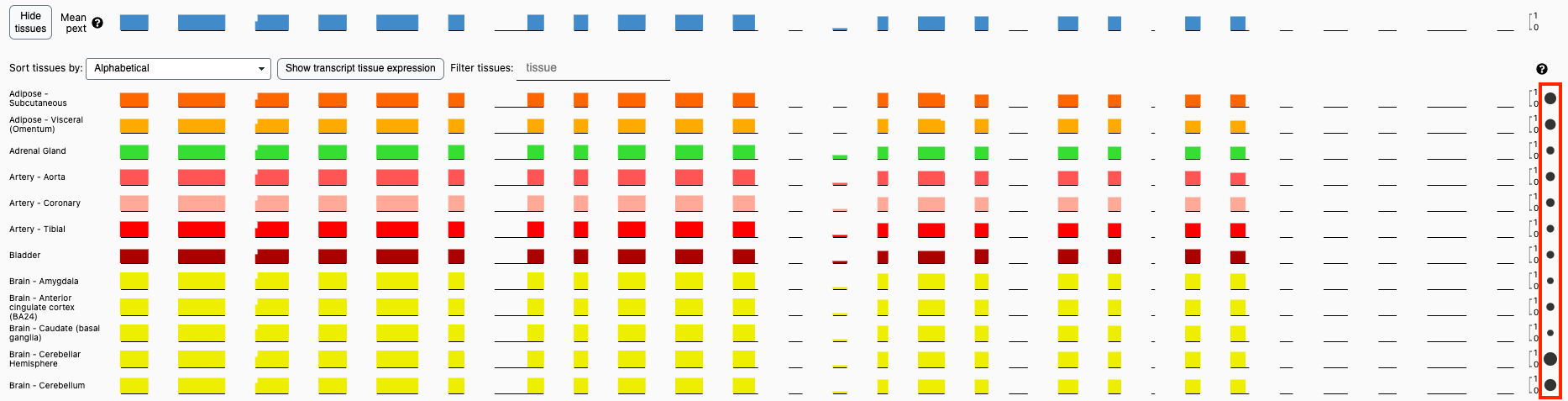
Now, the pext track shows a dot next to each tissue, the size of which is scaled based on the mean expression in that tissue across all the gene’s transcripts. Hovering the pointer over the dot shows the mean expression in that tissue across all transcripts as well as the transcript most expressed in that tissue.
The individual tissue pext tracks can also now be sorted by the mean transcript expression in the tissue.