We have released a new utility, the gnomAD toolbox, to enable easier analysis of gnomAD data. Community feedback has highlighted challenges with the usage of our downloadable files, which have become increasingly difficult to parse due to their growing size. As the gnomAD project continues to scale, we expect the size of these files to increase further. To ensure continued access to these data and empower all users, we have created a new open-source repository containing utility functions that allow users to query gnomAD data without requiring any data downloads.
Downloading the full gnomAD short variant release is not viable for all users due to the space required to store the data (~1.4 TB total for the v4 exomes and genomes VCFs!) and the computational resources needed to process such a large dataset. Certain queries, such as determining the total number of singleton variants within the gnomAD dataset, are currently only answerable by wrangling our downloadable data. To make these analyses more accessible to all users, we have created a new utility GitHub repository called the gnomAD toolbox. After installing the repository and its required software dependencies, the toolbox enables analyses on a user’s local computer without requiring a local copy of gnomAD data or prior knowledge of cloud computing.
The gnomAD toolbox is a work in progress. Our initial implementation includes building functionality that calculates metrics on the short variant data requested by users via our forum or email, including:
-
How to explore various release files (variants, all sites AN, coverage, etc.)
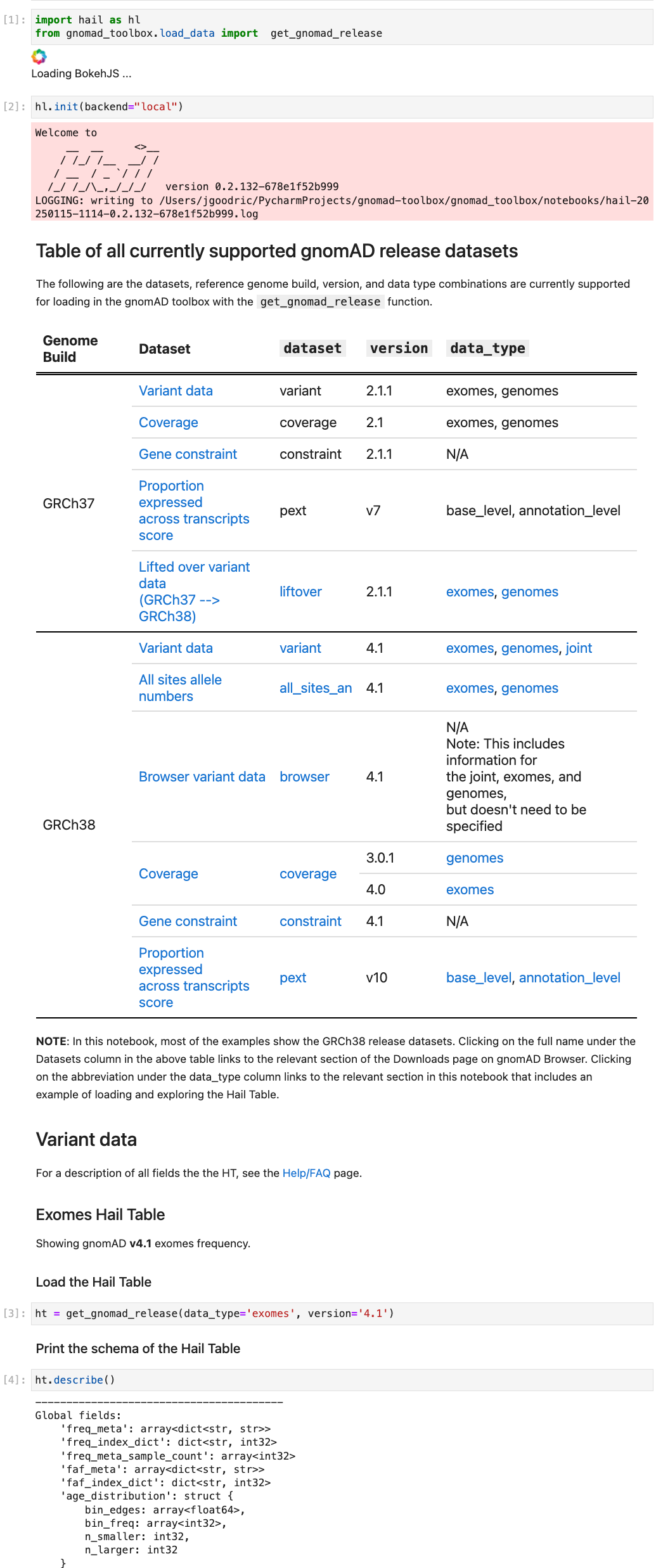
-
How to filter variants in a specific gene
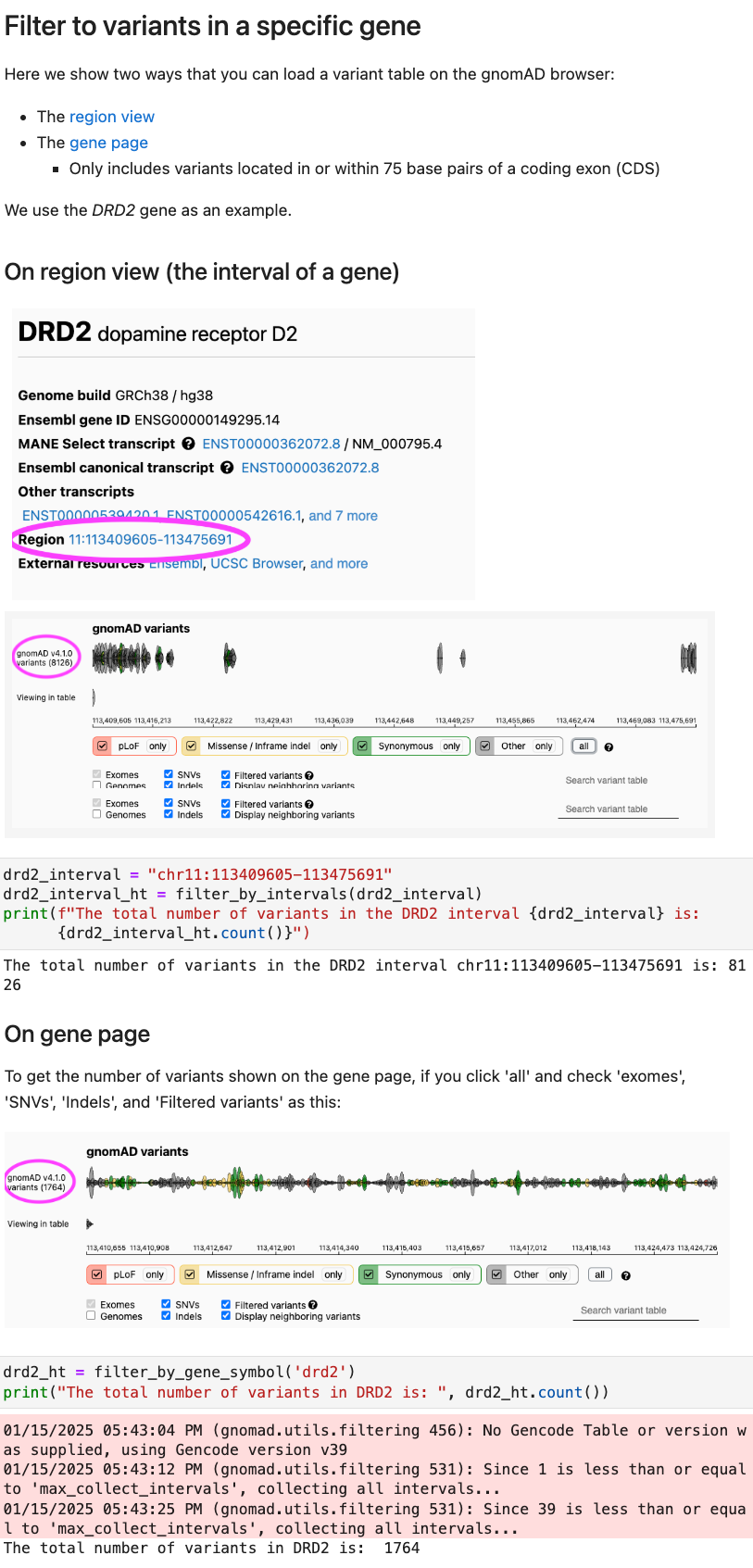
-
How to filter variants by VEP consequences
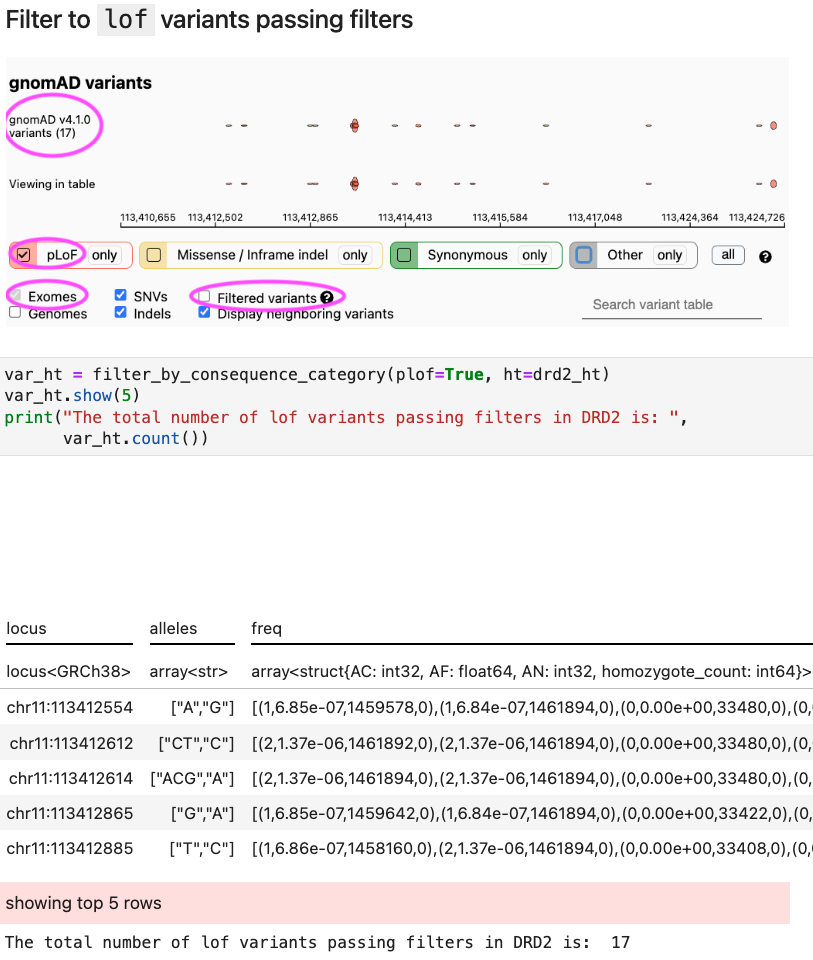
-
How to get the frequency information for specific genetic ancestry group(s)
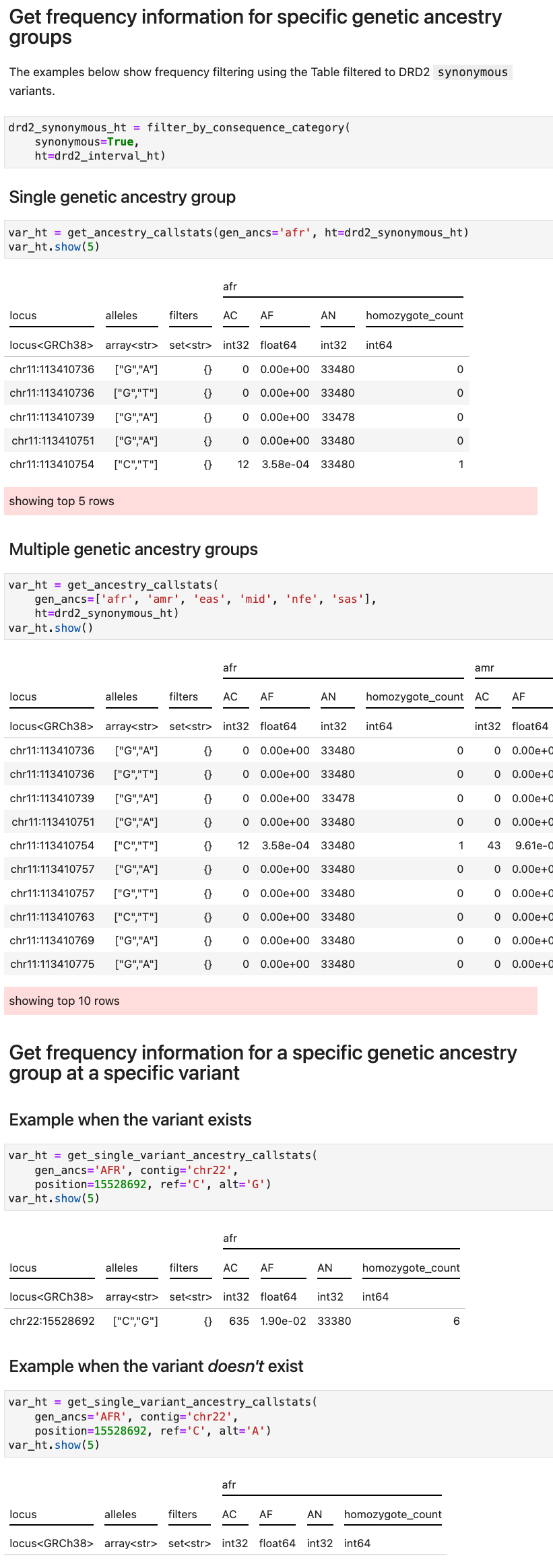
-
How to get variant counts by frequency bin

-
How to filter to predicted loss-of-function (pLoF) variants that we used to compute constraint metrics
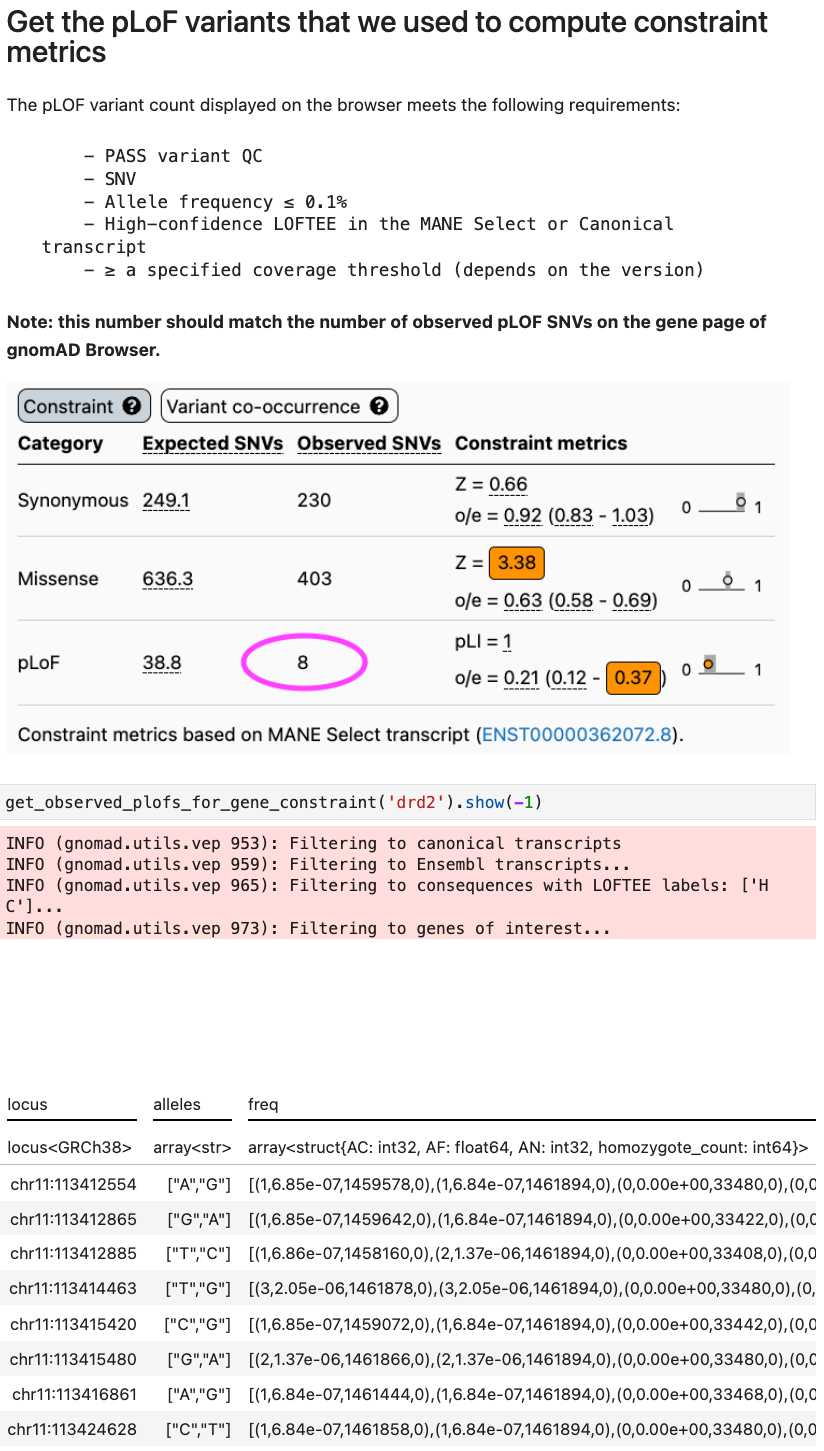
We are interested in crafting this toolbox with input from our community. We encourage our users to discuss any other desired functionality on our forum, and we highly encourage anyone interested in contributing new functionality to the toolbox to do so via pull request.
Acknowledgments
We thank the rest of the gnomAD Production team for brainstorming function ideas and specifically thank Michael Wilson for reviewing the blog post draft.
Updated in February 2025 to change author list ordering to reflect contributions to the gnomAD toolbox rather than blog post authorship.