This changelog entry discusses smaller updates to gnomAD added with the v4.1 release that were not discussed in the blog post.
As part of gnomAD v4.1, we have added the following new features:
- Browser display of non-UK Biobank (non-UKB) subset of gnomAD v4 exomes
- Supplementary files containing REVEL scores for variants in 2,414 transcripts that weren’t scored in the v4.0 release
- New joint exome + genome frequency resources with statistical tests flagging variants with discrepant frequencies
We have also updated the following features:
- Gene constraint metrics
- Exome copy number variants
- Genome structural variants
- Stats page
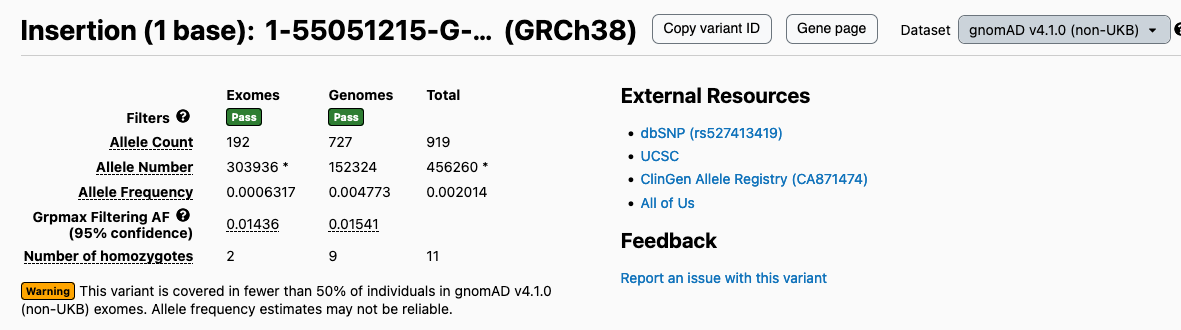
Browser display of non-UK Biobank subset
As part of gnomAD v4.1, we have exposed the non-UK Biobank (non-UKB) subset of the gnomAD v4 exomes. This subset was previously only available in our downloadable files but is now available in the “Dataset” toggle on gene, transcript, region, and variant pages. Note that this subset only impacts variant frequencies in the exomes; none of the genome samples were from the UK Biobank. Therefore, when the non-UKB subset is selected, the exome variant frequencies will update to reflect counts from the non-UKB subset, and the genome numbers will remain unchanged.
Example variant:
Supplementary REVEL files
We used a combination of transcript information (MANE Select or canonical) from Ensembl v105 and variant information (locus and alleles combination) to ascertain REVEL scores for variants in gnomAD v4.0 and v4.1 (exomes and genomes). However, REVEL was computed using Ensembl v64. This means that variants within 2,414 MANE Select transcripts in gnomAD v4.0 and v4.1 are missing REVEL scores because they were present in Ensembl v105 but not in Ensembl v64.
To address this, we annotated the variants within the 2,414 genes with the maximum REVEL score found at the specific locus and allele, rather than the score for the MANE Select or canonical transcript. We have released two TSVs; the exomes TSV adds REVEL scores to 1,936,321 out of 2,284,296 (87.77%) missense variants within the 2,414 genes. The genomes TSV adds REVEL scores to 528,204 out of 620,799 (85.08%) missense variants within the 2,414 genes.
Joint frequency VCFs
We have released a downloadable Hail Table and VCFs containing statistical tests that flag variants with discordant frequencies in the gnomAD exomes and genomes. For more information about these tests, please see our blog post and help page.
Gene constraint metrics
We regenerated the gene constraint metrics using gnomAD v4.1 frequencies and have updated the constraint table on the gene and transcript pages. Note that these updated metrics have the same caveats as the v4.0 metrics; for more details, please see our blog post.
Exome copy number variants
The exome copy number variants (CNVs) have been updated to include:
- Variant 193076__DUP relabeled as 16p12.2_DUP
- Refined annotation of 15q11 and 22q11 loci
- Updated N_EXN_VAR and N_INT_VAR labels for size of events to be consistent with the genic annotation used by short variant team
- Updated sex karyotype terminology (“XX” and “XY” instead of “Male” and “Female”)
- SC (site count), SN (site number), and SF (site frequency) labels reformatted to match the short variant VCF format (e.g., updating “afr_SC” to “SC_afr”)
Genome structural variants
The genome structural variants (SVs) have been updated to include:
- Genotype information for multiallelic CNVs (mCNVs)
- Updated genetic ancestry (e.g., “Other” to “Remaining”) and sex karyotype terminology (same as CNVs above)
- Updated gene annotations (using the same gene list used for the short variants)
- Updated metric annotation order to match the short variant VCF format (e.g., updating “nfe_AF” to “AF_nfe”)